class: center, middle  ### Advanced Machine Learning with scikit-learn # Imbalanced Data Andreas C. Müller Columbia University, scikit-learn .smaller[https://github.com/amueller/ml-training-advanced] ??? FIXME add AP for all evaluations, add more roc / pr curves FIXME better illustration for sampling FIXME better motivation for sampling / why we want to change things FIXME TOO SHORT FIXME Add estimators minimizing loss directly? --- class: spacious # Two sources of imbalance - Asymmetric cost - Asymmetric data ??? --- class: spacious # Why do we care? - Why should cost be symmetric? - All data is imbalanced - Detect rare events ??? --- # Changing Thresholds .tiny-code[ ```python data = load_breast_cancer() X_train, X_test, y_train, y_test = train_test_split( data.data, data.target, stratify=data.target, random_state=0) lr = LogisticRegression().fit(X_train, y_train) y_pred = lr.predict(X_test) classification_report(y_test, y_pred) ``` ``` precision recall f1-score support 0 0.91 0.92 0.92 53 1 0.96 0.94 0.95 90 avg/total 0.94 0.94 0.94 143 ``` ```python y_pred = lr.predict_proba(X_test)[:, 1] > .85 classification_report(y_test, y_pred) ``` ``` precision recall f1-score support 0 0.84 1.00 0.91 53 1 1.00 0.89 0.94 90 avg/total 0.94 0.93 0.93 143 ``` ] ??? --- # Roc Curve .center[  ] ??? --- class: center, middle ## Remedies for the model ??? --- class: compact # Mammography Data .smaller[ .center[  ] ```python X.shape ``` (11183, 6) ```python np.bincount(y) ``` array([10923, 260]) ] ??? Note: Split one slide to two due to size constraints. --- # Mammography Data .smaller[ ```python from sklearn.model_selection import cross_validate from sklearn.linear_model import LogisticRegression scores = cross_validate(LogisticRegression(), X_train, y_train, cv=10, scoring=('roc_auc', 'average_precision')) scores['test_roc_auc'].mean(), scores['test_average_precision'].mean() ``` 0.920, 0.630 ```python from sklearn.ensemble import RandomForestClassifier scores = cross_validate(RandomForestClassifier(n_estimators=100), X_train, y_train, cv=10, scoring=('roc_auc', 'average_precision')) scores['test_roc_auc'].mean(), scores['test_average_precision'].mean() ``` 0.939, 0.722 ] ??? --- # Basic Approaches .left-column[ .center[  ] ] .right-column[ </br> </br> </br> Change the training procedure ] ??? --- class: center, spacious # Sckit-learn vs resampling  ??? - The transform method only transforms X - Pipelines work by chaining transforms - To resample the data, we need to also change y --- class: spacious # Imbalance-Learn http://imbalanced-learn.org ``` pip install -U imbalanced-learn ``` Extends `sklearn` API --- ### Sampler To resample a data sets, each sampler implements: ```python data_resampled, targets_resampled = obj.sample(data, targets) ``` Fitting and sampling can also be done in one step: ```python data_resampled, targets_resampled = obj.fit_sample(data, targets) ``` -- <br /> <br /> In Pipelines: Sampling only done in `fit`! ??? - Imbalance-learn extends scikit-learn interface with a “sample” method. - Imbalance-learn has a custom pipeline that allows resampling. - Imbalance-learn: resampling is only performed during fitting - Warning: not everything in imbalance-learn is multiclass! --- # Random Undersampling ```python from imblearn.under_sampling import RandomUnderSampler rus = RandomUnderSampler(replacement=False) X_train_subsample, y_train_subsample = rus.fit_sample( X_train, y_train) print(X_train.shape) print(X_train_subsample.shape) print(np.bincount(y_train_subsample)) ``` ``` (8387, 6) (390, 6) [195 195] ``` ??? - Drop data from the majority class randomly - Often untill balanced - Very fast training (data shrinks to 2x minority) - Loses data ! --- # Random Undersampling .smaller[ ```python from imblearn.pipeline import make_pipeline as make_imb_pipeline undersample_pipe = make_imb_pipeline(RandomUnderSampler(), LogisticRegressionCV()) scores = cross_validate(undersample_pipe, X_train, y_train, cv=10, scoring=('roc_auc', 'average_precision')) scores['test_roc_auc'].mean(), scores['test_average_precision'].mean() # was 0.920, 0.630 without ``` ``` 0.927, 0.527 ``` ] -- .smaller[ ```python undersample_pipe_rf = make_imb_pipeline(RandomUnderSampler(), RandomForestClassifier()) scores = cross_validate(undersample_pipe_rf, X_train, y_train, cv=10, scoring=('roc_auc', 'average_precision')) scores['test_roc_auc'].mean(), scores['test_average_precision'].mean() # was 0.939, 0.722 without ``` ``` 0.951, 0.629 ``` ] ??? - As accurate with fraction of samples! - Really good for large datasets --- # Random Oversampling ```python from imblearn.over_sampling import RandomOverSampler ros = RandomOverSampler() X_train_oversample, y_train_oversample = ros.fit_sample( X_train, y_train) print(X_train.shape) print(X_train_oversample.shape) print(np.bincount(y_train_oversample)) ``` ``` (8387, 6) (16384, 6) [8192 8192] ``` ??? - Repeat samples from the minority class randomly - Often untill balanced - Much slower (dataset grows to 2x majority) --- # Random Oversampling .smaller[ ```python oversample_pipe = make_imb_pipeline(RandomOverSampler(), LogisticRegression()) scores = cross_validate(oversample_pipe, X_train, y_train, cv=10, scoring=('roc_auc', 'average_precision')) scores['test_roc_auc'].mean(), scores['test_average_precision'].mean() ``` 0.917, 0.585 ] -- .smaller[ ```python oversample_pipe_rf = make_imb_pipeline(RandomOverSampler(), RandomForestClassifier()) scores = cross_validate(oversample_pipe_rf, X_train, y_train, cv=10, scoring=('roc_auc', 'average_precision')) scores['test_roc_auc'].mean(), scores['test_average_precision'].mean() ``` 0.926, 0.715 ] ??? Logreg the same, Random Forest much worse than undersampling (about same as doing nothing) --- # Curves for LogReg .center[ 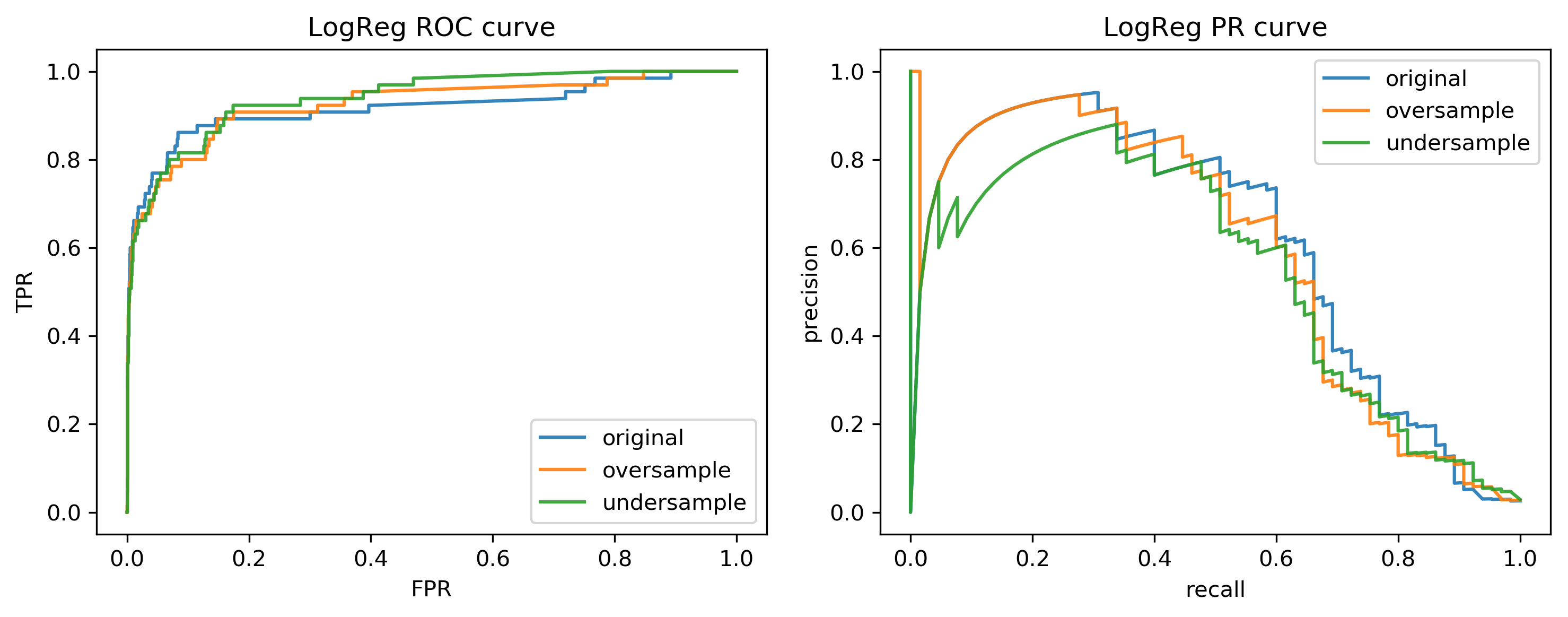 ] ??? --- # Curves for Random Forest .center[ 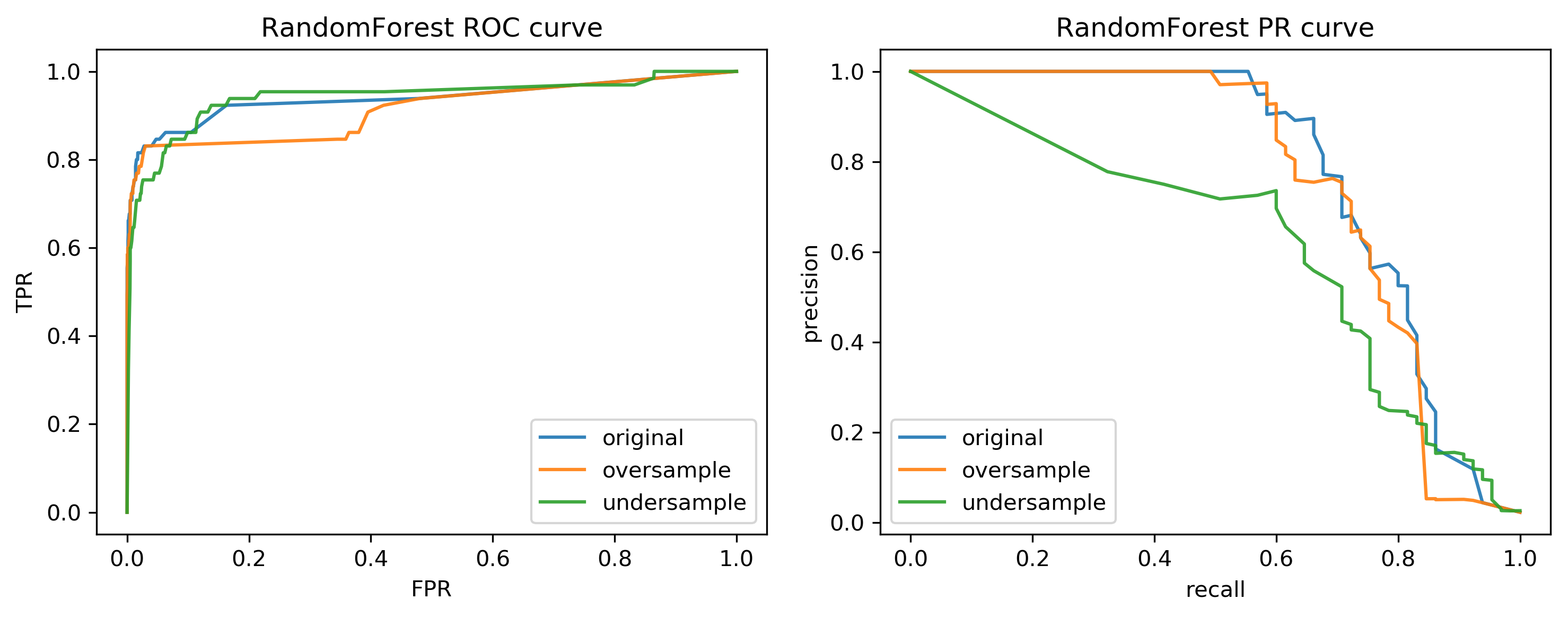 ] ??? --- # ROC or PR? FPR or Precision? `$$ \large\text{FPR} = \frac{\text{FP}}{\text{FP}+\text{TN}}$$` `$$ \large\text{Precision} = \frac{\text{TP}}{\text{TP}+\text{FP}}$$` ??? fIXME think of better explanation --- class: spacious # Class-weights - Instead of repeating samples, re-weight the loss function. - Works for most models! - Same effect as over-sampling (though not random), but not as expensive (dataset size the same). ??? --- class: spacious # Class-weights in linear models `$$\min_{w \in ℝ^{p}}-C \sum_{i=1}^n\log(\exp(-y_iw^T \textbf{x}_i) + 1) + ||w||_2^2$$` `$$ \min_{w \in \mathbb{R}^p} -C \sum_{i=1}^n c_{y_i} \log(\exp(-y_i w^T \mathbf{x}_i) + 1) + ||w||^2_2 $$` Similar for linear and non-linear SVM ??? --- # Class weights in trees Gini Index: `$$H_\text{gini}(X_m) = \sum_{k\in\mathcal{Y}} p_{mk} (1 - p_{mk})$$` `$$H_\text{gini}(X_m) = \sum_{k\in\mathcal{Y}} c_k p_{mk} (1 - p_{mk})$$` Prediction: Weighted vote ??? --- #Using Class-Weights .smaller[ ```python scores = cross_validate(LogisticRegression(class_weight='balanced'), X_train, y_train, cv=10, scoring=('roc_auc', 'average_precision')) scores['test_roc_auc'].mean(), scores['test_average_precision'].mean() ``` 0.918, 0.587 ```python scores = cross_validate(RandomForestClassifier(n_estimators=100, class_weight='balanced'), X_train, y_train, cv=10, scoring=('roc_auc', 'average_precision')) scores['test_roc_auc'].mean(), scores['test_average_precision'].mean() ``` 0.917, 0.701 ] ??? --- class: spacious # Ensemble Resampling - Random resampling separate for each instance in an ensemble! - Paper: “Exploratory Undersampling for Class Imbalance Learning” - Not in sklearn (yet) - Easy with imblearn ??? --- # Easy Ensemble with imblearn .smaller[ ```python from sklearn.tree import DecisionTreeClassifier from imblearn.ensemble import BalancedBaggingClassifier tree = DecisionTreeClassifier(max_features='auto') resampled_rf = BalancedBaggingClassifier(base_estimator=tree, n_estimators=100, random_state=0) scores = cross_validate(resampled_rf, X_train, y_train, cv=10, scoring=('roc_auc', 'average_precision')) scores['test_roc_auc'].mean(), scores['test_average_precision'].mean() ``` 0.957, 0.654 ] ??? -As cheap as undersampling, but much better results than anything else! </br> -Didn't do anything for Logistic Regression. --- class: center, middle 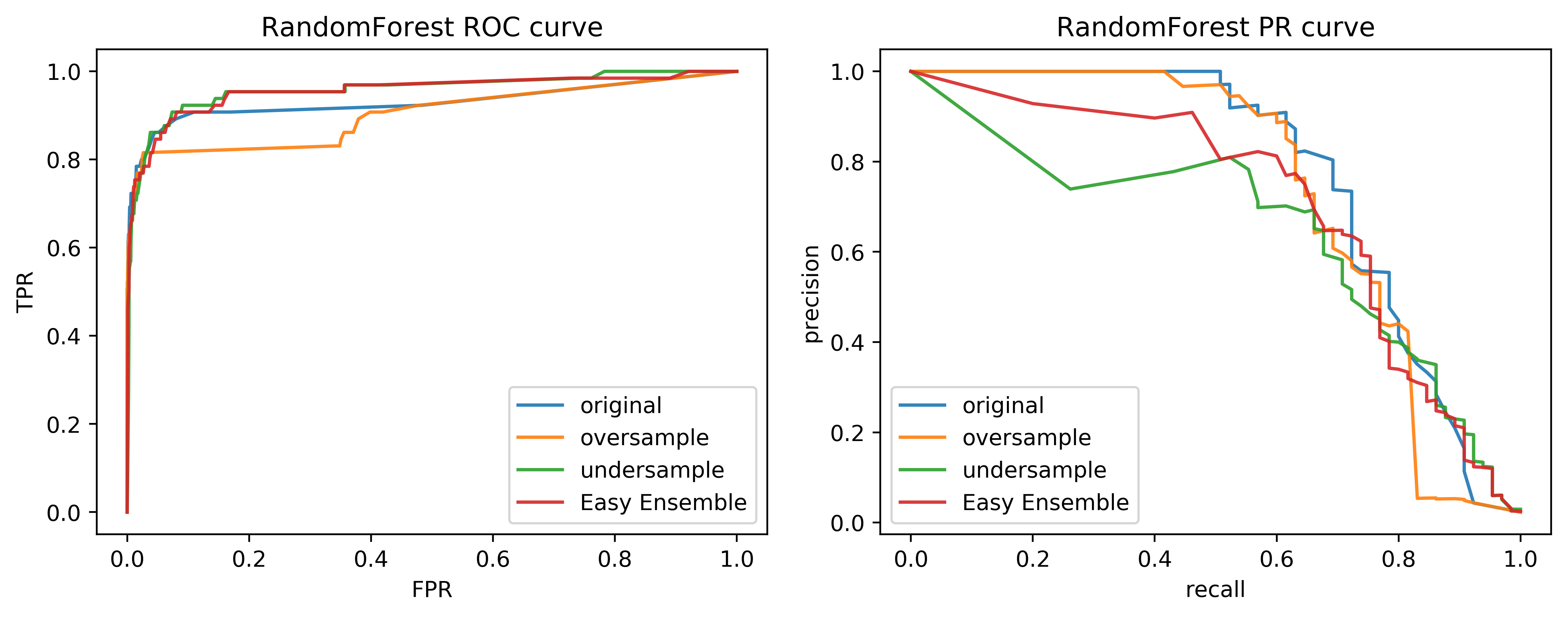 --- class: spacious # Summary - Always check roc_auc and AP look at curves - Undersampling is very fast and can help! - Undersampling + Ensembles is very powerful! - Can add synthetic samples with SMOTE ???